Welcome to PhyloToAST documentation!¶
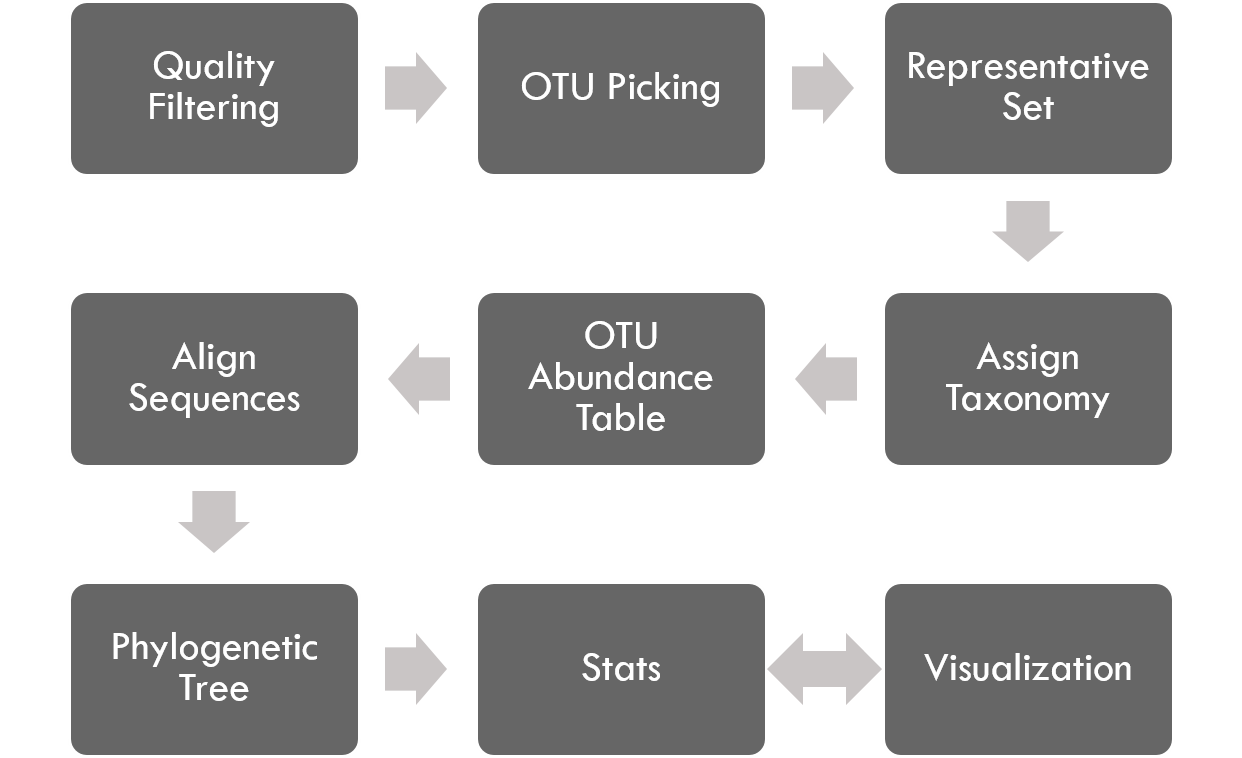
The PhyloToAST project is a collection of python scripts that modifies the original QIIME [1] pipeline:
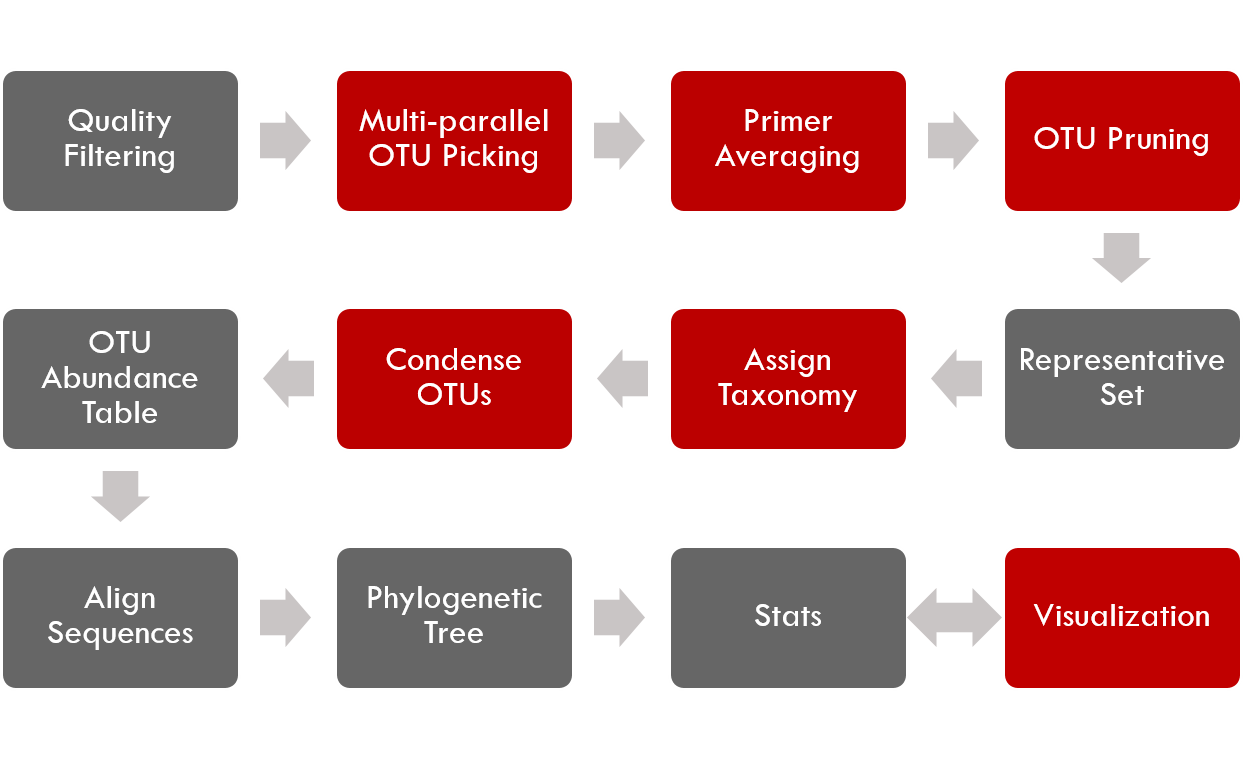
by adding or modifying several steps (as seen below) including support for PBS-based cluster-computing, multiple primer support [2], enhanced support for species-specific analysis, and additional visualization tools.
Download and install PhyloToAST package.
PhyloToAST Scripts¶
Citations:
| [1] | QIIME allows analysis of high-throughput community sequencing data. J Gregory Caporaso, Justin Kuczynski, Jesse Stombaugh, Kyle Bittinger, Frederic D Bushman, Elizabeth K Costello, Noah Fierer, Antonio Gonzalez Pena, Julia K Goodrich, Jeffrey I Gordon, Gavin A Huttley, Scott T Kelley, Dan Knights, Jeremy E Koenig, Ruth E Ley, Catherine A Lozupone, Daniel McDonald, Brian D Muegge, Meg Pirrung, Jens Reeder, Joel R Sevinsky, Peter J Turnbaugh, William A Walters, Jeremy Widmann, Tanya Yatsunenko, Jesse Zaneveld and Rob Knight; Nature Methods, 2010; doi: 10.1038/nmeth.f.303 |
| [2] | Target Region Selection Is a Critical Determinant of Community Fingerprints Generated by 16S Pyrosequencing. Kumar PS, Brooker MR, Dowd SE, Camerlengo T (2011) Target Region Selection Is a Critical Determinant of Community Fingerprints Generated by 16S Pyrosequencing. PLoS ONE 6(6): e20956. doi: 10.1371/journal.pone.0020956 |