PCoA_bubble.py¶
Create a series of Principal Coordinate plots for each OTU in an input list where the plot points are varied in size by the relative abundance of the OTU (relative to either Sample or the total contribution of the OTU to the data set.
usage: PCoA_bubble.py [-h] -i OTU_TABLE -m MAPPING -pc PCOA_FP -b GROUP_BY [-c COLORS] -ids OTU_IDS_FP [-o OUTPUT_DIR] [-s SAVE_AS] [--scale_by SCALE_BY] [--ggplot2_style] [-v]
Required Arguments¶
-
-iOTU_TABLE,--otu_tableOTU_TABLE¶ The biom-format file with OTU-Sample abundance data.
-
-mMAPPING,--mappingMAPPING¶ The mapping file specifying group information for each sample.
-
-pcPCOA_FP,--pcoa_fpPCOA_FP¶ Principal Coordinates Analysis file. Eg. unweighted_unifrac_pc.txt, or any other output from principal_coordinates.py.
-
-bGROUP_BY,--group_byGROUP_BY¶ Column name in mapping file specifying group information.
-
-cCOLORS,--colorsCOLORS¶ A column name in the mapping file containing hexadecimal (#FF0000) color values that will be used to color the groups. Each sample ID must have a color entry.
-
-idsOTU_IDS_FP,--otu_ids_fpOTU_IDS_FP¶ Path to a file containing one OTU ID per line. One plot will be created for each OTU.
Optional Arguments¶
-
-oOUTPUT_DIR,--output_dirOUTPUT_DIR¶ The directory to output the PCoA plots to.
-
-sSAVE_AS,--save_asSAVE_AS¶ The type of image file for PCoA plots. By default, files will be saved in SVG format.
-
--scale_bySCALE_BY¶ Species relative abundance is multiplied by this factor in order to make appropriate visible bubbles in the output plots. Default is 1000.
-
--ggplot2_style¶ Apply ggplot2 styling to the figure.
-
-v,--verbose¶ Displays species name as each is being plotted and saved.
-
-h,--help¶ Show this help message and exit
Example plot¶
PCoA bubble plots of subgingival microbiome pathogens of smokers [1].
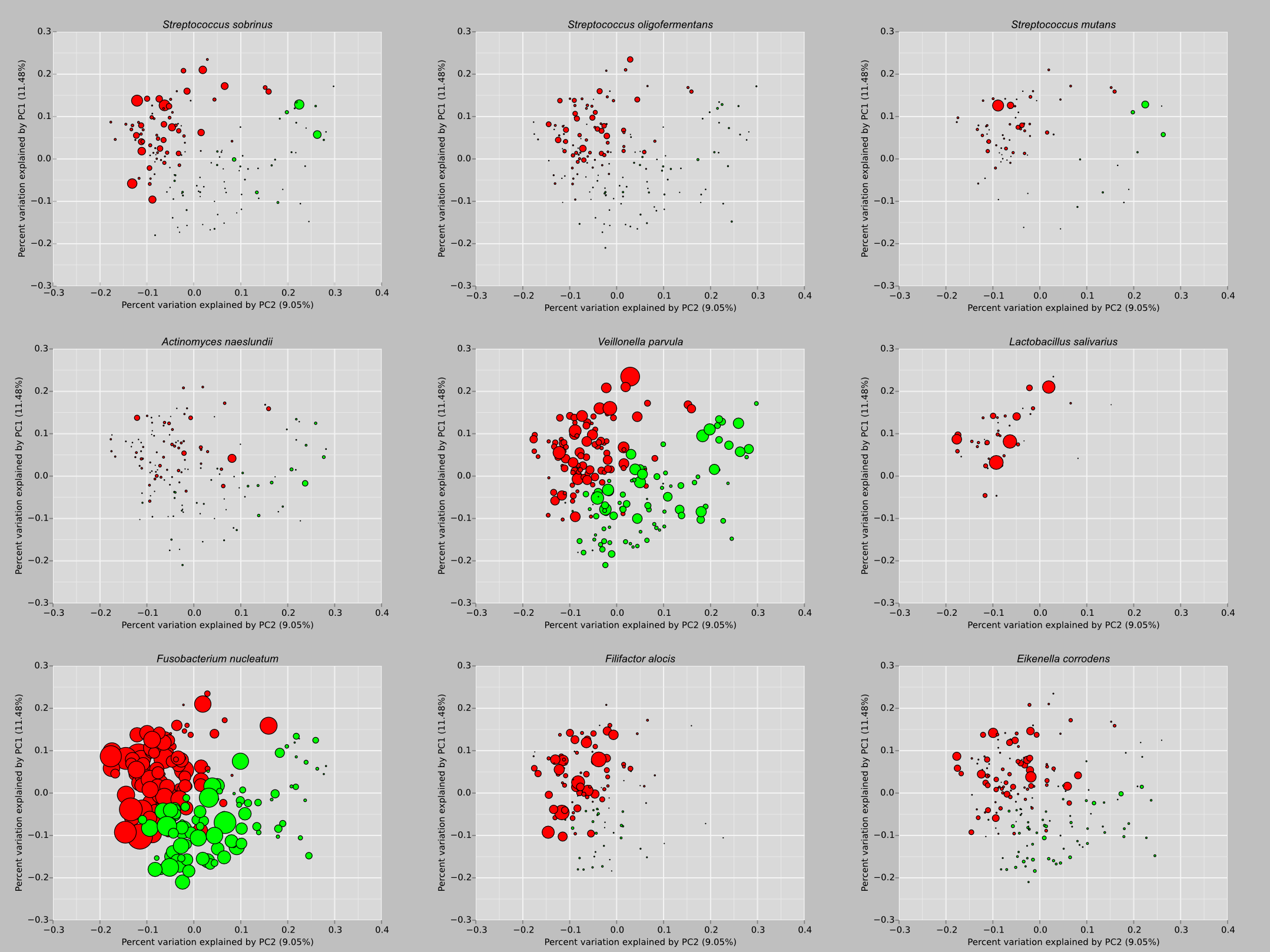
Citation:
| [1] | The Subgingival Microbiome of Clinically Healthy Current and Never Smokers. Matthew R Mason, Philip M Preshaw, Haikady N Nagaraja, Shareef M Dabdoub, Anis Rahman and Purnima S Kumar; doi: 10.1038/ismej.2014.114 |